Integrating National Ecological Observatory Network (NEON) Airborne Remote Sensing and In-Situ Data for Optimal Tree Species Classification
Abstract
:1. Introduction
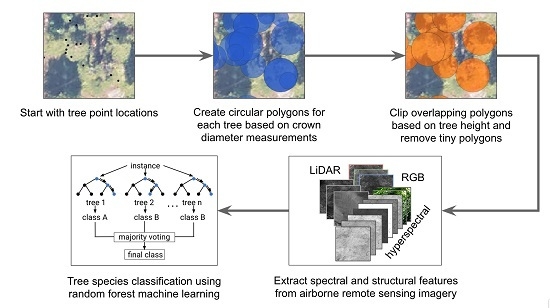
- Evaluate which training set preparation approach yields the most accurate tree species classification accuracy. We expected smaller tree polygons would capture more valuable variation in canopy features than using stem location points, and capture less noise and neighboring materials than larger circular polygons.
- Evaluate the value added of our proposed tree crown polygon clipping workflow, which removes tree crown polygons with small area values and clips overlapping tree crown regions based on associated in-situ tree height measurements.
- Assess the tree species classification accuracies achievable for the four dominant subalpine conifer species in a region of the Southern Rockies, Colorado, USA using the proposed NEON training data preparation approaches.
- Determine which NEON AOP imagery-derived features are the most important for predicting tree species to help inform overarching tree species classification efforts. We anticipated the hyperspectral imagery to be the most important compared to RGB or LiDAR-derived features.
- Contribute open reproducible tools so that the NEON data user community can use and build upon these techniques across diverse vegetated ecosystems.
2. Materials and Methods
2.1. Study Area and Field Data
2.2. Airborne Remote Sensing Data
2.3. Proposed Reference Data Preprocessing
2.4. Remote Sensing Feature Extraction
2.5. Random Forest Classification
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NEON | National Ecological Observatory Network |
| AOP | NEON’s Airborne Observation Platform |
| NIWO | Niwot Ridge Mountain Research Station NEON site |
| LiDAR | Light Detection and Ranging |
| CHM | Canopy Height Model |
| NIS | NEON Imaging Spectrometer |
| RGB | Red, Green, Blue multispectral imagery |
| RF | Random Forest classification |
References
- Martin, M.; Newman, S.; Aber, J.; Congalton, R. Determining forest species composition using high spectral resolution remote sensing data. Remote Sens. Environ. 1998, 65, 249–254. [Google Scholar] [CrossRef]
- Anderson, J.E.; Plourde, L.C.; Martin, M.E.; Braswell, B.H.; Smith, M.L.; Dubayah, R.O.; Hofton, M.A.; Blair, J.B. Integrating waveform lidar with hyperspectral imagery for inventory of a northern temperate forest. Remote Sens. Environ. 2008, 112, 1856–1870. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Fusion of hyperspectral and LIDAR remote sensing data for classification of complex forest areas. IEEE Trans. Geosci. Remote Sens. 2008, 46, 1416–1427. [Google Scholar] [CrossRef] [Green Version]
- Asner, G.P.; Martin, R.E.; Knapp, D.; Tupayachi, R.; Anderson, C.; Sinca, F.; Vaughn, N.; Llactayo, W. Airborne laser-guided imaging spectroscopy to map forest trait diversity and guide conservation. Science 2017, 355, 385–389. [Google Scholar] [CrossRef]
- Kaartinen, H.; Hyyppä, J.; Yu, X.; Vastaranta, M.; Hyyppä, H.; Kukko, A.; Holopainen, M.; Heipke, C.; Hirschmugl, M.; Morsdorf, F.; et al. An international comparison of individual tree detection and extraction using airborne laser scanning. Remote Sens. 2012, 4, 950–974. [Google Scholar] [CrossRef] [Green Version]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Wang, K.; Wang, T.; Liu, X. A review: Individual tree species classification using integrated airborne LiDAR and optical imagery with a focus on the urban environment. Forests 2019, 10, 1. [Google Scholar] [CrossRef] [Green Version]
- Hansen, M.C.; Potapov, P.V.; Moore, R.; Hancher, M.; Turubanova, S.; Tyukavina, A.; Thau, D.; Stehman, S.; Goetz, S.; Loveland, T.R.; et al. High-resolution global maps of 21st-century forest cover change. Science 2013, 342, 850–853. [Google Scholar] [CrossRef] [Green Version]
- Stavros, E.N.; Schimel, D.; Pavlick, R.; Serbin, S.; Swann, A.; Duncanson, L.; Fisher, J.B.; Fassnacht, F.; Ustin, S.; Dubayah, R.; et al. ISS observations offer insights into plant function. Nat. Ecol. Evol. 2017, 1, 0194. [Google Scholar] [CrossRef]
- He, Y.; Yang, J.; Caspersen, J.; Jones, T. An Operational Workflow of Deciduous-Dominated Forest Species Classification: Crown Delineation, Gap Elimination, and Object-Based Classification. Remote Sens. 2019, 11, 2078. [Google Scholar] [CrossRef] [Green Version]
- Anderson, K.; Gaston, K.J. Lightweight unmanned aerial vehicles will revolutionize spatial ecology. Front. Ecol. Environ. 2013, 11, 138–146. [Google Scholar] [CrossRef] [Green Version]
- Sankey, T.; Donager, J.; McVay, J.; Sankey, J.B. UAV lidar and hyperspectral fusion for forest monitoring in the southwestern USA. Remote Sens. Environ. 2017, 195, 30–43. [Google Scholar] [CrossRef]
- Nezami, S.; Khoramshahi, E.; Nevalainen, O.; Pölönen, I.; Honkavaara, E. Tree Species Classification of Drone Hyperspectral and RGB Imagery with Deep Learning Convolutional Neural Networks. Remote Sens. 2020, 12, 1070. [Google Scholar] [CrossRef] [Green Version]
- Asner, G.P.; Knapp, D.E.; Boardman, J.; Green, R.O.; Kennedy-Bowdoin, T.; Eastwood, M.; Martin, R.E.; Anderson, C.; Field, C.B. Carnegie Airborne Observatory-2: Increasing science data dimensionality via high-fidelity multi-sensor fusion. Remote Sens. Environ. 2012, 124, 454–465. [Google Scholar] [CrossRef]
- Liu, L.; Coops, N.C.; Aven, N.W.; Pang, Y. Mapping urban tree species using integrated airborne hyperspectral and LiDAR remote sensing data. Remote Sens. Environ. 2017, 200, 170–182. [Google Scholar] [CrossRef]
- Shen, X.; Cao, L. Tree-species classification in subtropical forests using airborne hyperspectral and LiDAR data. Remote Sens. 2017, 9, 1180. [Google Scholar] [CrossRef] [Green Version]
- Krzystek, P.; Serebryanyk, A.; Schnörr, C.; Červenka, J.; Heurich, M. Large-Scale Mapping of Tree Species and Dead Trees in Šumava National Park and Bavarian Forest National Park Using Lidar and Multispectral Imagery. Remote Sens. 2020, 12, 661. [Google Scholar] [CrossRef] [Green Version]
- Wietecha, M.; Jełowicki, Ł.; Mitelsztedt, K.; Miścicki, S.; Stereńczak, K. The capability of species-related forest stand characteristics determination with the use of hyperspectral data. Remote Sens. Environ. 2019, 231, 111232. [Google Scholar] [CrossRef]
- Lin, Y.; Hyyppä, J. A comprehensive but efficient framework of proposing and validating feature parameters from airborne LiDAR data for tree species classification. Int. J. Appl. Earth Obs. Geoinf. 2016, 46, 45–55. [Google Scholar] [CrossRef]
- Naidoo, L.; Cho, M.A.; Mathieu, R.; Asner, G. Classification of savanna tree species, in the Greater Kruger National Park region, by integrating hyperspectral and LiDAR data in a Random Forest data mining environment. ISPRS J. Photogramm. Remote Sens. 2012, 69, 167–179. [Google Scholar] [CrossRef]
- Yu, X.; Hyyppä, J.; Litkey, P.; Kaartinen, H.; Vastaranta, M.; Holopainen, M. Single-sensor solution to tree species classification using multispectral airborne laser scanning. Remote Sens. 2017, 9, 108. [Google Scholar] [CrossRef] [Green Version]
- Budei, B.C.; St-Onge, B.; Hopkinson, C.; Audet, F.A. Identifying the genus or species of individual trees using a three-wavelength airborne lidar system. Remote Sens. Environ. 2018, 204, 632–647. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree species classification with random forest using very high spatial resolution 8-band WorldView-2 satellite data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef] [Green Version]
- Ballanti, L.; Blesius, L.; Hines, E.; Kruse, B. Tree species classification using hyperspectral imagery: A comparison of two classifiers. Remote Sens. 2016, 8, 445. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Zhang, X. Object-Based Tree Species Classification Using Airborne Hyperspectral Images and LiDAR Data. Forests 2020, 11, 32. [Google Scholar] [CrossRef] [Green Version]
- Hauglin, M.; Ørka, H.O. Discriminating between Native Norway Spruce and Invasive Sitka Spruce—A comparison of multitemporal Landsat 8 imagery, aerial images and airborne laser scanner data. Remote Sens. 2016, 8, 363. [Google Scholar] [CrossRef] [Green Version]
- Osińska-Skotak, K.; Radecka, A.; Piórkowski, H.; Michalska-Hejduk, D.; Kopeć, D.; Tokarska-Guzik, B.; Ostrowski, W.; Kania, A.; Niedzielko, J. Mapping Succession in Non-Forest Habitats by Means of Remote Sensing: Is the Data Acquisition Time Critical for Species Discrimination? Remote Sens. 2019, 11, 2629. [Google Scholar] [CrossRef] [Green Version]
- Weinstein, B.G.; Marconi, S.; Bohlman, S.; Zare, A.; White, E. Individual tree-crown detection in RGB imagery using semi-supervised deep learning neural networks. Remote Sens. 2019, 11, 1309. [Google Scholar] [CrossRef] [Green Version]
- Pal, M. Random forest classifier for remote sensing classification. Int. J. Remote Sens. 2005, 26, 217–222. [Google Scholar] [CrossRef]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Koch, B. A framework for mapping tree species combining hyperspectral and LiDAR data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Piiroinen, R.; Heiskanen, J.; Maeda, E.; Viinikka, A.; Pellikka, P. Classification of tree species in a diverse African agroforestry landscape using imaging spectroscopy and laser scanning. Remote Sens. 2017, 9, 875. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.X.; Tuia, D.; Mou, L.; Xia, G.S.; Zhang, L.; Xu, F.; Fraundorfer, F. Deep learning in remote sensing: A comprehensive review and list of resources. IEEE Geosci. Remote Sens. Mag. 2017, 5, 8–36. [Google Scholar] [CrossRef] [Green Version]
- Maxwell, A.E.; Warner, T.A.; Fang, F. Implementation of machine-learning classification in remote sensing: An applied review. Int. J. Remote Sens. 2018, 39, 2784–2817. [Google Scholar] [CrossRef] [Green Version]
- Fricker, G.A.; Ventura, J.D.; Wolf, J.A.; North, M.P.; Davis, F.W.; Franklin, J. A convolutional neural network classifier identifies tree species in mixed-conifer forest from hyperspectral imagery. Remote Sens. 2019, 11, 2326. [Google Scholar] [CrossRef] [Green Version]
- Franklin, S.; Hall, R.; Moskal, L.; Maudie, A.; Lavigne, M. Incorporating texture into classification of forest species composition from airborne multispectral images. Int. J. Remote Sens. 2000, 21, 61–79. [Google Scholar] [CrossRef]
- Maschler, J.; Atzberger, C.; Immitzer, M. Individual tree crown segmentation and classification of 13 tree species using airborne hyperspectral data. Remote Sens. 2018, 10, 1218. [Google Scholar] [CrossRef] [Green Version]
- Marconi, S.; Graves, S.J.; Gong, D.; Nia, M.S.; Le Bras, M.; Dorr, B.J.; Fontana, P.; Gearhart, J.; Greenberg, C.; Harris, D.J.; et al. A data science challenge for converting airborne remote sensing data into ecological information. PeerJ 2019, 6, e5843. [Google Scholar] [CrossRef] [Green Version]
- Ghiyamat, A.; Shafri, H.Z.M.; Shariff, A.R.M. Influence of tree species complexity on discrimination performance of vegetation Indices. Eur. J. Remote Sens. 2016, 49, 15–37. [Google Scholar] [CrossRef] [Green Version]
- Michener, W.K. Ecological data sharing. Ecol. Inform. 2015, 29, 33–44. [Google Scholar] [CrossRef] [Green Version]
- Keller, M.; Schimel, D.S.; Hargrove, W.W.; Hoffman, F.M. A continental strategy for the National Ecological Observatory Network. Front. Ecol. Environ. 2008, 6, 282–284. [Google Scholar] [CrossRef]
- Kampe, T.U.; Johnson, B.R.; Kuester, M.A.; Keller, M. NEON: The first continental-scale ecological observatory with airborne remote sensing of vegetation canopy biochemistry and structure. J. Appl. Remote Sens. 2010, 4, 043510. [Google Scholar] [CrossRef]
- Johnson, B.R.; Kuester, M.A.; Kampe, T.U.; Keller, M. National ecological observatory network (NEON) airborne remote measurements of vegetation canopy biochemistry and structure. In Proceedings of the 2010 IEEE International Geoscience and Remote Sensing Symposium, Honolulu, HI, USA, 25–30 July 2010; IEEE: Piscataway, NJ, USA, 2010; pp. 2079–2082. [Google Scholar]
- Kao, R.H.; Gibson, C.M.; Gallery, R.E.; Meier, C.L.; Barnett, D.T.; Docherty, K.M.; Blevins, K.K.; Travers, P.D.; Azuaje, E.; Springer, Y.P.; et al. NEON terrestrial field observations: Designing continental-scale, standardized sampling. Ecosphere 2012, 3, 1–17. [Google Scholar] [CrossRef]
- Balch, J.K.; Nagy, R.C.; Halpern, B.S. NEON is seeding the next revolution in ecology. Front. Ecol. Environ. 2020, 18, 3. [Google Scholar] [CrossRef] [Green Version]
- Hampton, S.E.; Jones, M.B.; Wasser, L.A.; Schildhauer, M.P.; Supp, S.R.; Brun, J.; Hernandez, R.R.; Boettiger, C.; Collins, S.L.; Gross, L.J.; et al. Skills and knowledge for data-intensive environmental research. BioScience 2017, 67, 546–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carpenter, J. May the Best Analyst Win. Science 2011, 331, 698–699. [Google Scholar] [CrossRef]
- Graves, S.; Gearhart, J.; Caughlin, T.T.; Bohlman, S. A digital mapping method for linking high-resolution remote sensing images to individual tree crowns. PeerJ Prepr. 2018, 6, e27182v1. [Google Scholar]
- National Ecological Observatory Network. Data Products NEON.DP1.10098.001, NEON.DP3.30006.001, NEON.DP3.30026.001, NEON.DP3.30015.001, NEON.DP3.30025.001, NEON.DP3.30010.001. 2019. Available online: http://data.neonscience.org (accessed on 31 January 2019).
- Bajorski, P. Statistical inference in PCA for hyperspectral images. IEEE J. Sel. Top. Signal Process. 2011, 5, 438–445. [Google Scholar] [CrossRef]
- Rouse, J.; Haas, R.; Schell, J.; Deering, D. Monitoring vegetation systems in the Great Plains with ERTS. NASA Spec. Publ. 1974, 351, 309. [Google Scholar]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Kaufman, Y.J.; Tanre, D. Atmospherically resistant vegetation index (ARVI) for EOS-MODIS. IEEE Trans. Geosci. Remote Sens. 1992, 30, 261–270. [Google Scholar] [CrossRef]
- Gamon, J.; Penuelas, J.; Field, C. A narrow-waveband spectral index that tracks diurnal changes in photosynthetic efficiency. Remote Sens. Environ. 1992, 41, 35–44. [Google Scholar] [CrossRef]
- Serrano, L.; Penuelas, J.; Ustin, S.L. Remote sensing of nitrogen and lignin in Mediterranean vegetation from AVIRIS data: Decomposing biochemical from structural signals. Remote Sens. Environ. 2002, 81, 355–364. [Google Scholar] [CrossRef]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Walsh, S.J. Coniferous tree species mapping using Landsat data. Remote Sens. Environ. 1980, 9, 11–26. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Congalton, R.G.; Green, K. Assessing the Accuracy Of Remotely Sensed Data: Principles and Practices; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Koch, B.; Kattenborn, T.; Straub, C.; Vauhkonen, J. Segmentation of forest to tree objects. In Forestry Applications of Airborne Laser Scanning; Springer: Berlin/Heidelberg, Germany, 2014; pp. 89–112. [Google Scholar]
- Weinstein, B.G.; Marconi, S.; Bohlman, S.A.; Zare, A.; White, E.P. Cross-site learning in deep learning RGB tree crown detection. Ecol. Inform. 2020, 56, 101061. [Google Scholar] [CrossRef]
- Alonzo, M.; Bookhagen, B.; Roberts, D.A. Urban tree species mapping using hyperspectral and lidar data fusion. Remote Sens. Environ. 2014, 148, 70–83. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Vescovo, L.; Wohlfahrt, G.; Balzarolo, M.; Pilloni, S.; Sottocornola, M.; Rodeghiero, M.; Gianelle, D. New spectral vegetation indices based on the near-infrared shoulder wavelengths for remote detection of grassland phytomass. Int. J. Remote Sens. 2012, 33, 2178–2195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jollineau, M.; Howarth, P. Mapping an inland wetland complex using hyperspectral imagery. Int. J. Remote Sens. 2008, 29, 3609–3631. [Google Scholar] [CrossRef]








| Scientific Name (Common Name) | Species Code | Number of Mapped Trees |
|---|---|---|
| Abies lasiocarpa (Subalpine fir) | ABLAL | 249 |
| Pinus contorta (Lodgepole pine) | PICOL | 112 |
| Picea engelmannii (Engelmann spruce) | PIEN | 264 |
| Pinus flexilis (Limber pine) | PIFL2 | 74 |
| Feature Name | Description | Inputs/Equations | Reference |
|---|---|---|---|
| PC1, PC2 | 1st and 2nd principal components | 426-band Hyperspectral reflectance (372 after removing bad bands due to atmospheric absorption) 381–2509 nm with 5 nm spacing | [49] |
| Hyperspectral reflectance bands: | |||
| Normalized Difference Vegetation Index (NDVI) | [50] | ||
| Enhanced Vegetation Index (EVI) | [51] | ||
| Atmospherically Resistant Vegetation Index (ARVI) | [52] | ||
| Vegetation Indices | Canopy Xanthophyll, or Photochemical Reflectance Index (PRI) | [53] | |
| Normalized Difference Lignin Index (NDLI) | [54] | ||
| Normalized Difference Nitrogen Index (NDNI) | [54] | ||
| Soil-Adjusted Vegetation Index (SAVI) | [55] | ||
| CHM | Height of canopy above the ground | LiDAR-derived Digital Surface Model (DSM) – Digital Terrain Model (DTM) with modified data pit filling algorithm | [20] |
| Slope | Steepness of bare earth surface | DTM bare earth elevation ratio: height over distance | [56] |
| Aspect | Compass direction of steepest slope | DTM bare earth elevation degrees clockwise from North | [56] |
| rgb_mean_sd_R | Mean plus standard | ||
| rgb_mean_sd_G | deviation of red, green, | RGB multispectral bands | [35] |
| rgb_mean_sd_B | and blue (RGB) image | ||
| intensity values |
| Training Set | OOB Accuracy | IV Accuracy | Kappa |
|---|---|---|---|
| Points | 0.682 | 0.458 | 0.538 |
| Polygons-half diameter | 0.690 | 0.597 | 0.573 |
| Polygons-max diameter | 0.624 | 0.590 | 0.490 |
| Points-half diam clipped | 0.598 | 0.528 | 0.434 |
| Polygons-half diam clipped | 0.693 | 0.604 | 0.578 |
| Polygons-max diam clipped | 0.645 | 0.611 | 0.516 |
| Predicted Species | ||||||
|---|---|---|---|---|---|---|
| ABLAL | PICOL | PIEN | PIFL2 | PA % | ||
| ABLAL | 28 | 18 | 31 | 9 | 32.6 | |
| PICOL | 4 | 110 | 38 | 11 | 67.5 | |
| True species | PIEN | 8 | 30 | 111 | 19 | 66.1 |
| PIFL2 | 1 | 2 | 5 | 149 | 94.9 | |
| UA % | 68.3 | 68.8 | 60.0 | 79.3 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scholl, V.M.; Cattau, M.E.; Joseph, M.B.; Balch, J.K. Integrating National Ecological Observatory Network (NEON) Airborne Remote Sensing and In-Situ Data for Optimal Tree Species Classification. Remote Sens. 2020, 12, 1414. https://doi.org/10.3390/rs12091414
Scholl VM, Cattau ME, Joseph MB, Balch JK. Integrating National Ecological Observatory Network (NEON) Airborne Remote Sensing and In-Situ Data for Optimal Tree Species Classification. Remote Sensing. 2020; 12(9):1414. https://doi.org/10.3390/rs12091414
Chicago/Turabian StyleScholl, Victoria M., Megan E. Cattau, Maxwell B. Joseph, and Jennifer K. Balch. 2020. "Integrating National Ecological Observatory Network (NEON) Airborne Remote Sensing and In-Situ Data for Optimal Tree Species Classification" Remote Sensing 12, no. 9: 1414. https://doi.org/10.3390/rs12091414